Exploratory data analysis¶
Based on assignment 3
This Demo and the attached files from course PSYO/NESC 3505 Neural Data Science, at Dalhousie University. You should not disseminate, distribute or copy.
I am NOT post inappropriate or copyrighted content, advertise or promote outside products or organizations
© Copyright 2020.PSYO/NESC 3505 FAll 2020 https://dalpsychneuro.github.io/NESC_3505_textbook/intro.html
For demonstration purposes only
Background¶
Flanker Data We’ll be working with data from a similar flanker experiment as we used in Assignment 2. However, in Assignment 2 we gave you a “sanitized” version of the data, where we had done a bit of clean-up to extract just the information we wanted you to work with. In this assignment you’ll get experience starting with the raw data and extracting the necessary information yourself.
–PSYO 3505 Fall 2019 Assignment 2 cell1
Read file and check it¶
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
First of all we need input and examine the data
subjects = ['001_2015_05_22_11_30',
'002_2015_05_25_14_36',
'003_2015_05_28_14_09'
]
in_file = subjects[0]+'/'+subjects[0]+'_data.txt'
#input to data_frame
df = pd.read_csv(in_file)
df.head(5)
| id\tyear\tmonth\tday\thour\tminute\tgender\tage\thandedness\twait\tblock\ttrial\ttarget_location\ttarget\tflankers\trt\tresponse\terror\tpre_target_response\tITI_response\ttarget_on_error | |
|---|---|
| 0 | 001\t2015\t05\t22\t11\t30\tm\t25\tr\t3.24\tpra... |
| 1 | 001\t2015\t05\t22\t11\t30\tm\t25\tr\t3.24\tpra... |
| 2 | 001\t2015\t05\t22\t11\t30\tm\t25\tr\t3.24\tpra... |
| 3 | 001\t2015\t05\t22\t11\t30\tm\t25\tr\t3.24\tpra... |
| 4 | 001\t2015\t05\t22\t11\t30\tm\t25\tr\t3.24\tpra... |
looks like not very well, we need add escape character. Typically, I prefer read csv file directly and then check the head.
df = pd.read_csv(in_file,sep = '\t')
df
| id | year | month | day | hour | minute | gender | age | handedness | wait | ... | trial | target_location | target | flankers | rt | response | error | pre_target_response | ITI_response | target_on_error | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.240 | ... | 1 | left | black | incongruent | NaN | NaN | NaN | False | False | 0.023 |
| 1 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.240 | ... | 2 | right | black | incongruent | 0.550 | black | True | False | False | 0.023 |
| 2 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.240 | ... | 3 | up | white | incongruent | 0.565 | white | True | False | False | 0.024 |
| 3 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.240 | ... | 4 | up | black | congruent | 0.453 | black | True | False | False | 0.024 |
| 4 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.240 | ... | 5 | down | black | congruent | 0.442 | black | True | False | False | 0.023 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 187 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 28 | left | white | congruent | 0.349 | white | True | False | False | 0.024 |
| 188 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 29 | right | white | congruent | 0.371 | white | True | False | False | 0.023 |
| 189 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 30 | up | black | incongruent | 0.549 | black | True | False | False | 0.023 |
| 190 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 31 | left | white | neutral | 0.463 | white | True | False | False | 0.023 |
| 191 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 32 | right | black | neutral | 0.430 | black | True | False | False | 0.023 |
192 rows × 21 columns
look at the head and tail again
df.head(5)
| id | year | month | day | hour | minute | gender | age | handedness | wait | ... | trial | target_location | target | flankers | rt | response | error | pre_target_response | ITI_response | target_on_error | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.24 | ... | 1 | left | black | incongruent | NaN | NaN | NaN | False | False | 0.023 |
| 1 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.24 | ... | 2 | right | black | incongruent | 0.550 | black | True | False | False | 0.023 |
| 2 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.24 | ... | 3 | up | white | incongruent | 0.565 | white | True | False | False | 0.024 |
| 3 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.24 | ... | 4 | up | black | congruent | 0.453 | black | True | False | False | 0.024 |
| 4 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 3.24 | ... | 5 | down | black | congruent | 0.442 | black | True | False | False | 0.023 |
5 rows × 21 columns
df.tail(5)
| id | year | month | day | hour | minute | gender | age | handedness | wait | ... | trial | target_location | target | flankers | rt | response | error | pre_target_response | ITI_response | target_on_error | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 187 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 28 | left | white | congruent | 0.349 | white | True | False | False | 0.024 |
| 188 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 29 | right | white | congruent | 0.371 | white | True | False | False | 0.023 |
| 189 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 30 | up | black | incongruent | 0.549 | black | True | False | False | 0.023 |
| 190 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 31 | left | white | neutral | 0.463 | white | True | False | False | 0.023 |
| 191 | 1 | 2015 | 5 | 22 | 11 | 30 | m | 25 | r | 1.627 | ... | 32 | right | black | neutral | 0.430 | black | True | False | False | 0.023 |
5 rows × 21 columns
Then we check have missing value
print(pd.isna(df).sum())
id 0
year 0
month 0
day 0
hour 0
minute 0
gender 0
age 0
handedness 0
wait 0
block 0
trial 0
target_location 0
target 0
flankers 0
rt 2
response 2
error 2
pre_target_response 0
ITI_response 0
target_on_error 0
dtype: int64
Data cleaning¶
Obviously, we need to remove missing data by appling
.dropna() method
df = df.dropna()
print(pd.isna(df).sum())
id 0
year 0
month 0
day 0
hour 0
minute 0
gender 0
age 0
handedness 0
wait 0
block 0
trial 0
target_location 0
target 0
flankers 0
rt 0
response 0
error 0
pre_target_response 0
ITI_response 0
target_on_error 0
dtype: int64
This experiment has ‘practice’, the main idea is giving participants a chance to figure out the experiment. But we do not need it in our EDA. Not all experiments have ‘practice’ .
value = df[‘block’] print(value.unique())
df = df[df.block!='practice']
value = df['block']
print(value.unique())
['1' '2' '3' '4' '5']
Then we found ‘rt’ column is not milliseconds and need to convert
df['rt']
32 0.528
33 0.419
34 0.514
35 0.406
36 0.502
...
187 0.349
188 0.371
189 0.549
190 0.463
191 0.430
Name: rt, Length: 160, dtype: float64
df['rt'] = df['rt']*1000
print(df['rt'])
32 528.0
33 419.0
34 514.0
35 406.0
36 502.0
...
187 349.0
188 371.0
189 549.0
190 463.0
191 430.0
Name: rt, Length: 160, dtype: float64
‘rt’ is our interested variable, but In most behavioural studies, RTs are not normally distributed, so we need to Examining the RT distribution
df['rt'].describe()
count 160.00000
mean 465.16875
std 101.47170
min 314.00000
25% 392.25000
50% 454.00000
75% 505.25000
max 845.00000
Name: rt, dtype: float64
plot a histogram of RTs
df['rt'].plot(kind='hist')
# add a solid line at the median and dashed lines at the 25th and 75th percentiles (done for you)
plt.axvline(df['rt'].describe()['25%'], 0, 1, color='turquoise', linestyle='--')
plt.axvline(df['rt'].median(), 0, 1, color='cyan', linestyle='-')
plt.axvline(df['rt'].describe()['75%'], 0, 1, color='turquoise', linestyle='--')
# Rememebr to use plt.show() to see your plots (often they show anyway, but with some garbagy text at the top)
plt.show()
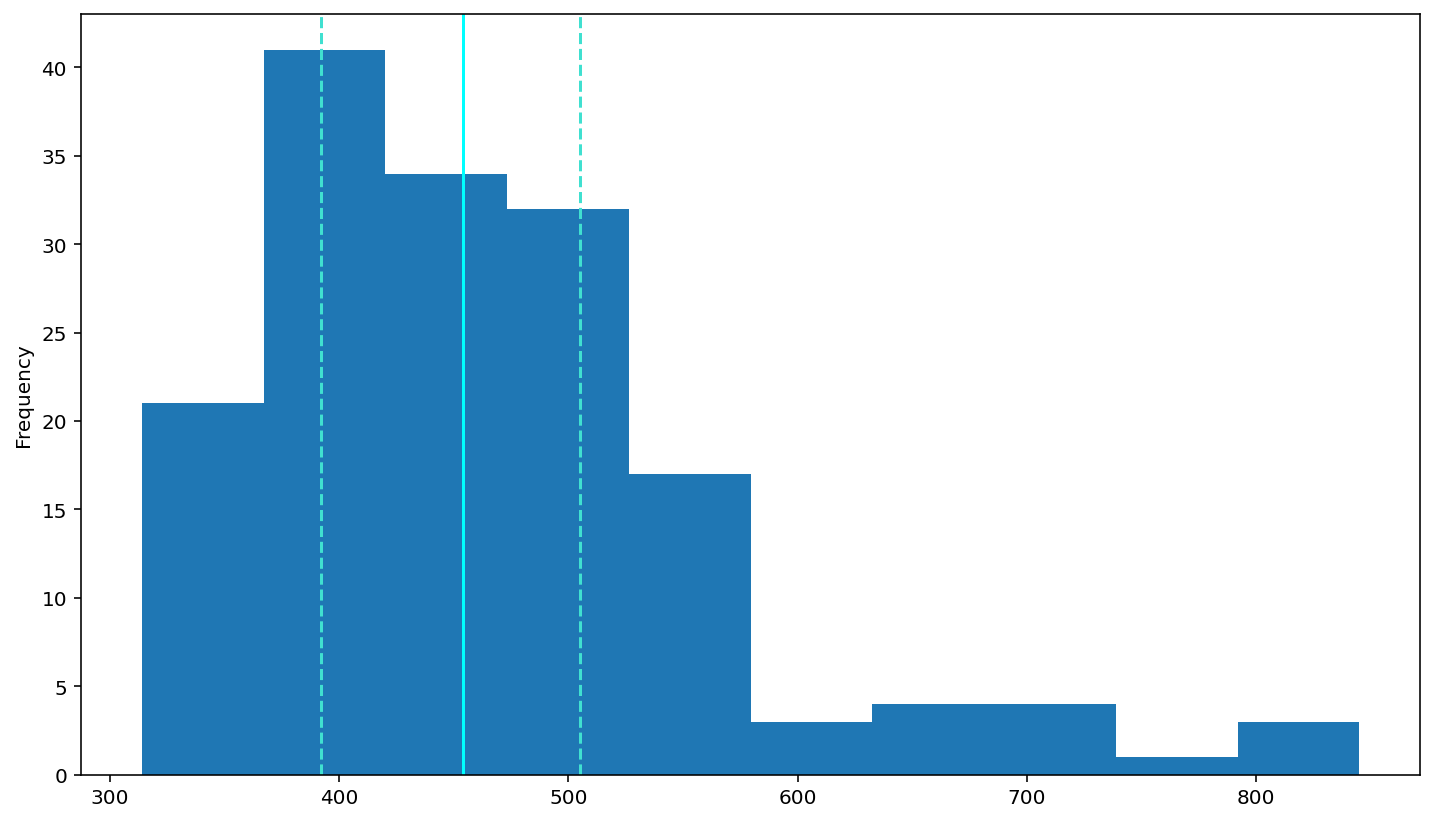
Analyzing relationships between variables¶
According to above figure, the histogram look not follow normal distribution, Looks like right skewed. Because the data sets have low ‘high value
check the cdf
df['rt'].plot(kind = 'hist',cumulative = True, density =True)
# add a dashed oragne line at the median
plt.axvline(df['rt'].describe()['25%'], 0, 1, color='turquoise', linestyle='--')
plt.axvline(df['rt'].median(), 0, 1, color='cyan', linestyle='-')
plt.axvline(df['rt'].describe()['75%'], 0, 1, color='turquoise', linestyle='--')
plt.show()
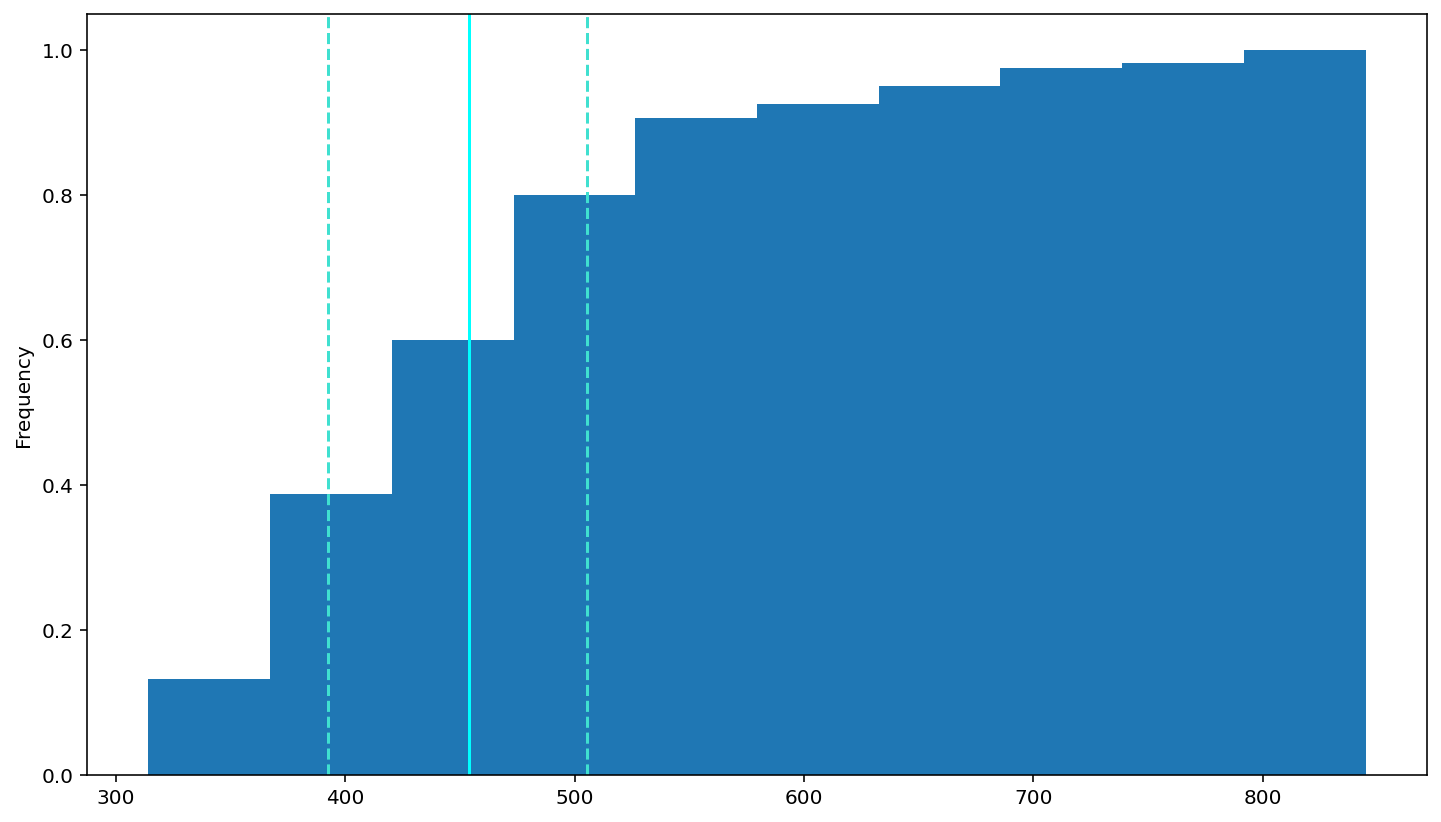
How should we do in this situation. The course tells me :
While the skew in the RT data makes sense, for the reasons described above, it’s problematic when running statistics on the data. This is because many conventional statistical tests, like t-tests and ANOVAs, assume that the data are normally distributed. Using skewed data can cause unreliable results.
For this reason, many researchers apply some transformation to RTs to make their distribution more normal (statistically normal, that is). A common one is to take the logarithm of the RT values: log(RT)log(RT); another is to take the inverse: 1/RT.
—PSYO 3505 Fall 2019 Assignment 2 cell40
So, for log(RT)
# log-transform the rt data (done for you)
df['logrt'] = np.log(df['rt'])
# put your plotting code here
# YOUR CODE HERE
df['logrt'].plot(kind='hist')
# add a solid line at the median and dashed lines at the 25th and 75th percentiles (done for you)
plt.axvline(df['logrt'].describe()['25%'], 0, 1, color='turquoise', linestyle='--')
plt.axvline(df['logrt'].median(), 0, 1, color='cyan', linestyle='-')
plt.axvline(df['logrt'].describe()['75%'], 0, 1, color='turquoise', linestyle='--')
# Rememebr to use plt.show() to see your plots (often they show anyway, but with some garbagy text at the top)
plt.show()
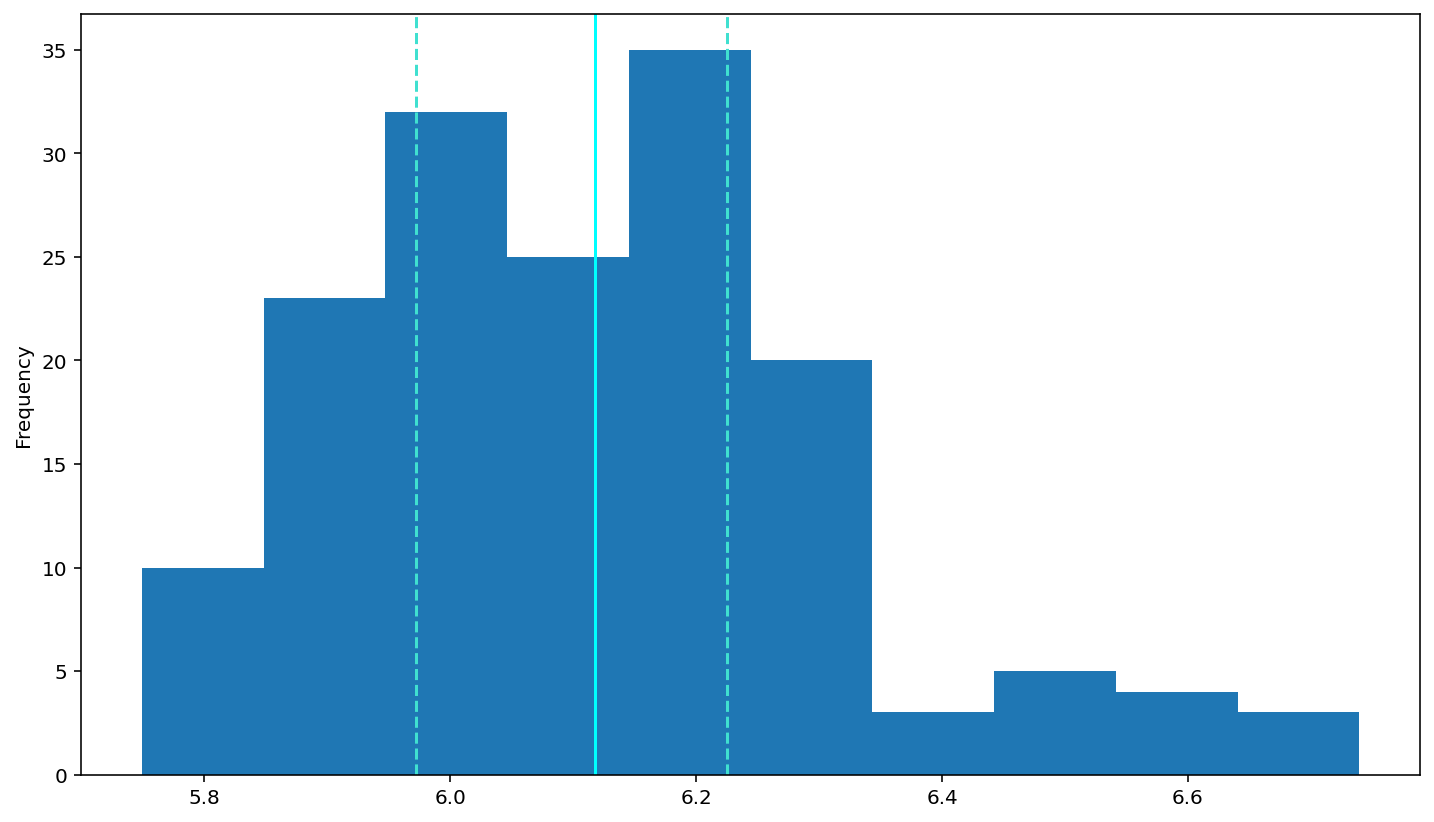
inverse: \(1/RT\)
df['invrt'] = 1/(df['rt'])
df['invrt'].plot(kind='hist')
# add a solid line at the median and dashed lines at the 25th and 75th percentiles (done for you)
plt.axvline(df['invrt'].describe()['25%'], 0, 1, color='turquoise', linestyle='--')
plt.axvline(df['invrt'].median(), 0, 1, color='cyan', linestyle='-')
plt.axvline(df['invrt'].describe()['75%'], 0, 1, color='turquoise', linestyle='--')
# Rememebr to use plt.show() to see your plots (often they show anyway, but with some garbagy text at the top)
plt.show()
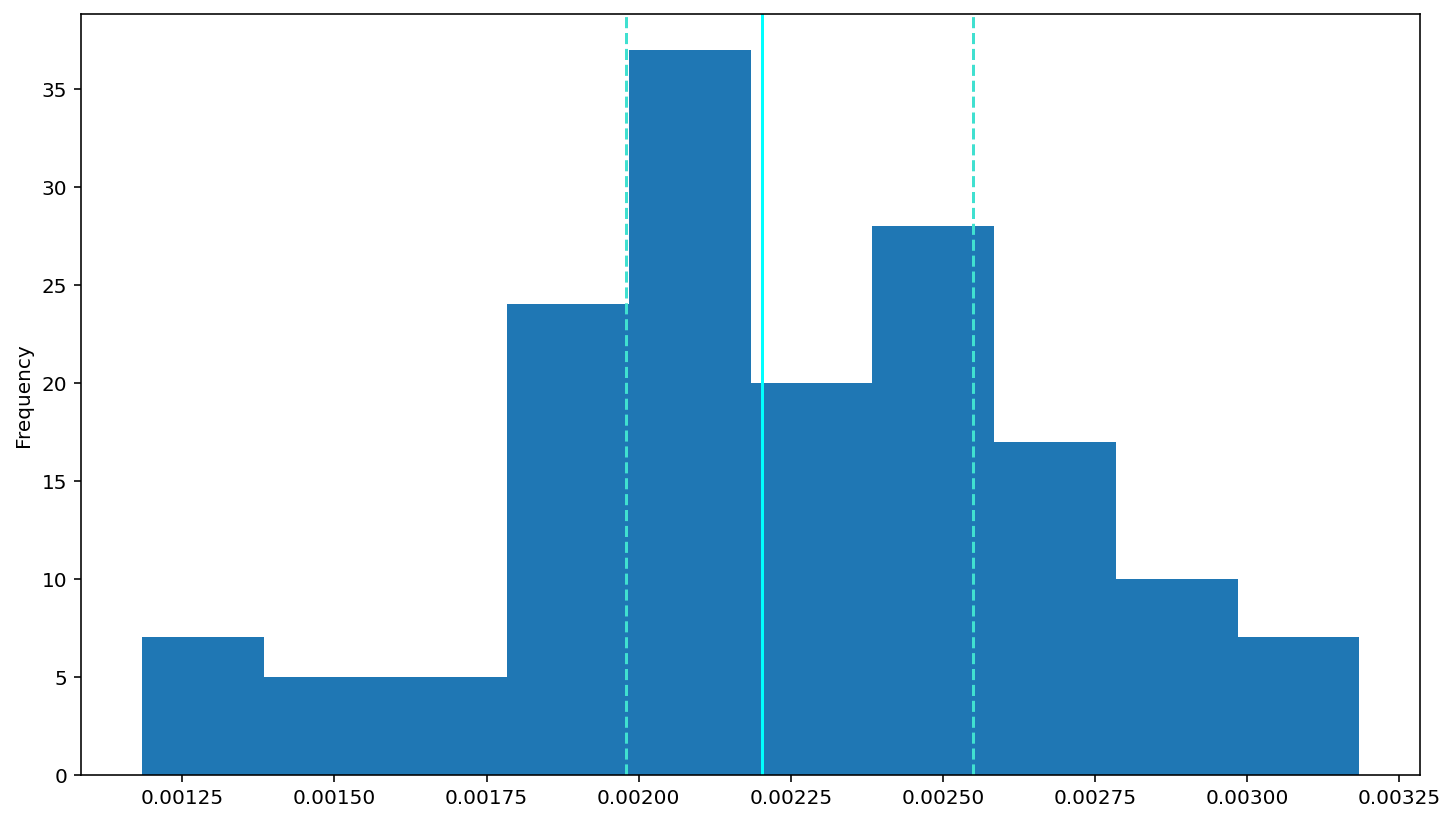
After that, we want to explore the data will be on errors and reaction times (RTs).
Recall that these data are from a “flanker” experiment in which participants had to respond with a left or right button press, depending on whether the target (centre) arrow pointed left or right. The target arrow was flanked with two arrows on either side that were either congruent (pointed in same direction) or incongruent (opposite direction).
—PSYO 3505 Fall 2019 Assignment 2 cell48
Tip
You can found more information about flanker experiment in this article ‘Flanker and Simon effects interact at the response selection stage’
rt_mean = df.groupby(by = 'flankers')[['rt','error']].agg('mean')
rt_mean = rt_mean.drop('neutral')
rt_mean
| rt | |
|---|---|
| flankers | |
| congruent | 452.60 |
| incongruent | 466.05 |
Generate a table for accuracy (the error column).
correct = df.groupby(by = 'flankers')[['error']].agg('sum')
correct.columns = ['Correct']
correct = correct.drop('neutral')
correct
| Correct | |
|---|---|
| flankers | |
| congruent | 40 |
| incongruent | 34 |
showing the accuracy rate
total= df.groupby(by = 'flankers')[['error']].agg('count')
correct = df.groupby(by = 'flankers')[['error']].agg('sum')
number = correct[:2]/total[:2]
number.columns = ['Accuracy']
number
| Accuracy | |
|---|---|
| flankers | |
| congruent | 1.00 |
| incongruent | 0.85 |
Visualization¶
Plot box plots of the log-transformed RT data (logrt) for each flanker conditio
boxplot1 = df.loc[df['flankers']=='congruent','logrt'].plot(kind = 'box',title = 'Congruent')
plt.show()
boxplot2 = df.loc[df['flankers']=='incongruent','logrt'].plot(kind = 'box',title = 'Incongruent')
plt.show()
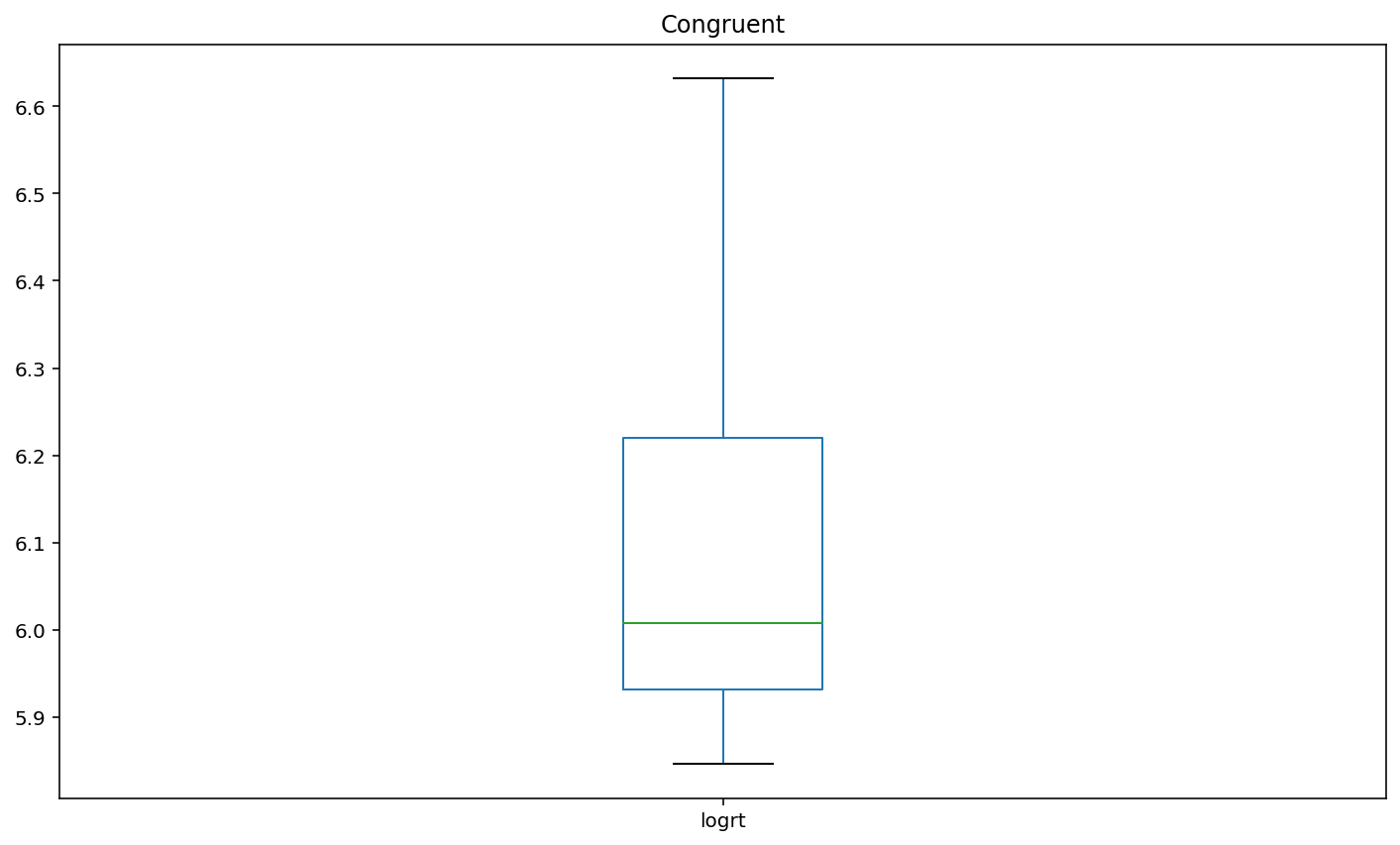
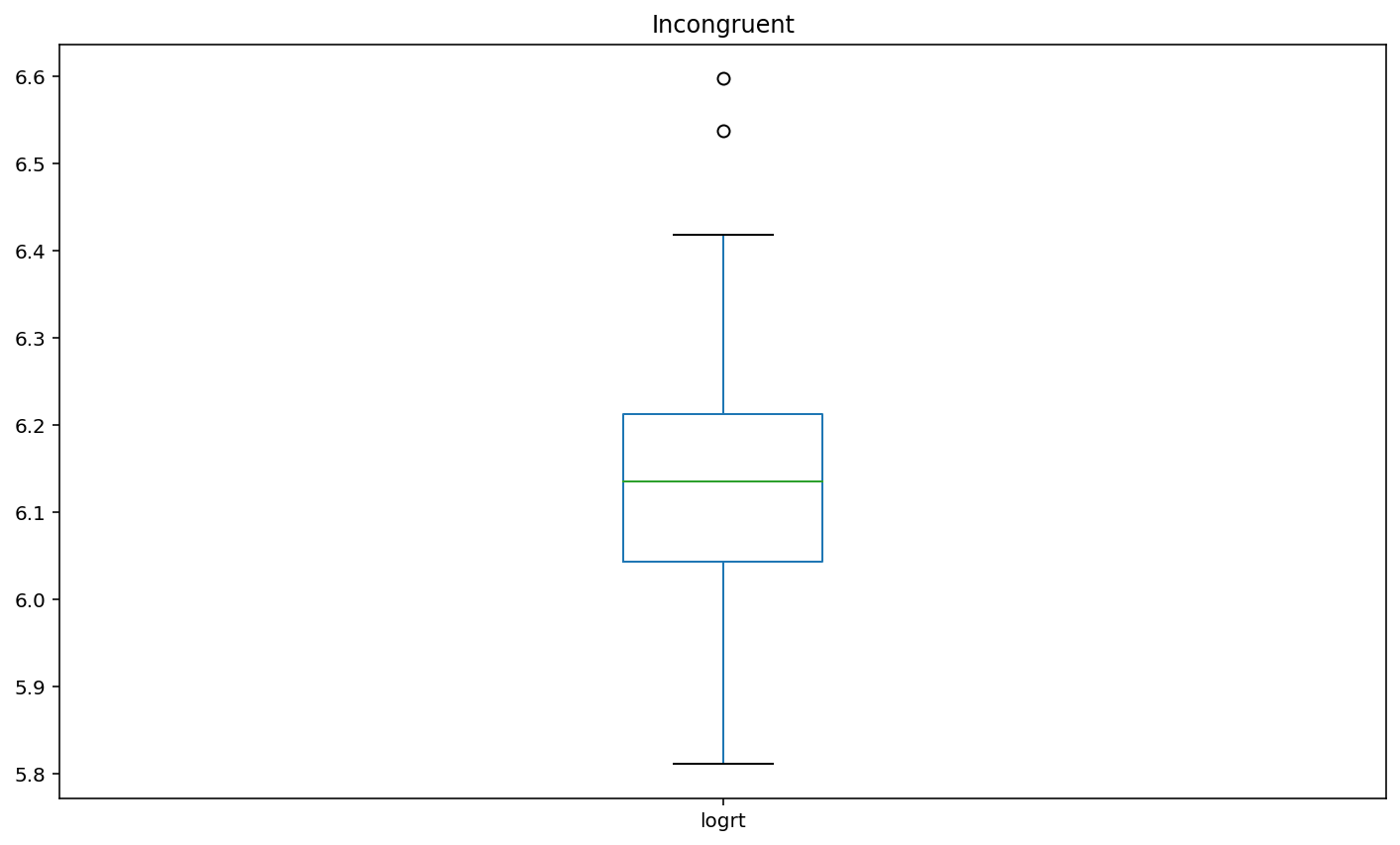
Interpretation¶
Basis of EDA, I can say incongruent flankers would lead to slower and less accurate responses. according to the participant results, the congruent condition has 100% accuracy and 452.60 (mean) response time. Incongruent condition has 85% accuracy and 466.05 (mean) response time.
So far, we end EDA methods (basic).
Through this assignment, I learned the basic steps of data analysis. Especially,EDA methods:
Read files
Data cleaning
Analyzing relationships between variables
Visualization
Interpretation
References¶
[1] NESC 3505 Neural Data Science, at Dalhousie University. Textbook
[2] NESC 3505 Neural Data Science, at Dalhousie University. Assignment 3
[3] An Extensive Step by Step Guide to Exploratory Data Analysis
Based on assignment 3
This Demo and the attached files from course PSYO/NESC 3505 Neural Data Science, at Dalhousie University. You should not disseminate, distribute or copy.
I am NOT post inappropriate or copyrighted content, advertise or promote outside products or organizations
© Copyright 2020.PSYO/NESC 3505 FAll 2020 https://dalpsychneuro.github.io/NESC_3505_textbook/intro.html
For demonstration purposes only